Research
Drug characterization by MALDI imaging mass spectrometry
Today:
Optimise lung retention and correlate drug PK in lung with effect and toxicity when only total concentration of drug in lung tissue homogenate can be measured.
Business benefit of tissue imaging:
Localisation of unlabelled drugs and metabolites and peptides/proteins in tissue
PK/PD ? does compound distribute to target site?
Toxicity ? does compound distribute specifically to affected tissue/cell type?
Drive chemistry towards compounds with optimal distribution ? FIC & BIC
Applicable to Lung/Liver/Kidney/Brain Pathophysiology
PSA is the major diagnostic marker for prostate cancer; however, it is not fully reliable. Therefore, it is of great interest to collect more information about PSA, including the identification of its possible variants,isoforms and phenotypes. By employing modern mass spectrometry based proteome analysis it is achievable to collect detailed comprehensive data on PSA composition in large number of samples. This work holds a promise to find significant novelty about PSA that allows us to understand why it can be useful diagnostic marker in many cases but irrelevant in other cases.
We will use sophisticated high resolution mass spectrometry techniques, both LC-ESI-MS/MS and MALDI-MS/MS, in order to collect complementary information about PSA. LC-ESI-MS and MALDI-MS methods will be established in order to analyze PSA peptides, fragmented with trypsin. Following method development screening of clinical samples will be performed. Posttranslational modifications of PSA will be extensively investigated. Statistical evaluation of the resulted data will be also performed.
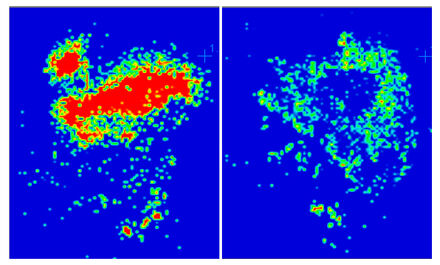
We are involved in Lung cancer research developments, to discover proteins related to disease initiation as well as proteins that relate to drug treatment – like personalized treatment with “Drug responder” patients. Cancer disease is the leading diagnosed cancer as well as the leading cause of cancer death globally.
We run studies with the objective that accounts for 13 % (1.82 million) of the total new cancer cases and 18 % (1.59 million) of the deaths in cancer in 2012 around the world. The five year survival for all stage lung cancer patients is only 49 about 15 %, and for stage IV patients.
Chromosome aberrations related to lung oncogenesis mechanisms have been revealed recently. About 50 % of the lung adenocarcinoma (ADC) tumors bear “driver mutations”. EGFR mutations are the most common drive mutations in lung ADC, usually due to losses in exon 19, or point mutations of exon 21 in chromosome 7.
Our Chromosome 19 Project (C-HPP) is a global consortium dedicated to mapping the entire human complement of proteins, with global membership (
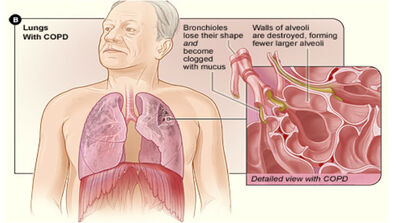
What is COPD:
In COPD, less air flows in and out of the airways because of;
- The walls of the airways become thick and inflamed;
- The airways make more mucus than usual, which can clog them;
- The airways and air sacs lose their elastic quality; and
- The walls between air sacs are destroyed (emphysema)
After many years of smoking both smokers and former smokers are at risk from developing smoking related diseases such as COPD, cardiovascular disease and lung cancer. In collaboration with an open clinic in Malmö (Sweden) we have an ongoing clinical study that is unique to the majority of other registered COPD studies. The study is longitudinal over five years looking for prognostic biomarkers in patients with early signs of COPD in smokers and former smokers.
Novel biomarkers may be discovered and subsequently implemented in clinical routine diagnostics for patients at risk from or already diagnosed with COPD and related diseases.
KOL-Örestad study will give the material for biomarker development. We collect clinical demographics from 200 smokers and ex-smokers with COPD to be compared with 50 healthy never smokers and 50 healthy smokers (age 35 to 80 years old). The enrolled patients go through a health examination including medical history, smoking history, lung function measurements, quality of life questionnaire and blood samples are drawn every six months during five years. The study has been approved by the regional ethical review board in Lund, (Approval number: DNR 213/480) and is registered at the NIH clinical trial registry (ClinTril.gov).
Blood samples will be analyzed using immunochemical techniques and mass spectrometry.
We expect to find protein and peptides that will be either up or down regulated and closely associated with disease status. Today there are 145 subjects enrolled and 30 have completed three visits.
The research in our group within the BAGADILICO environment focuses on Parkinson’s disease (PD), Alzheimer´s disease (AD) and Huntington’s disease (HD) biomarkers.
Our research group is working on the development of mass spectrometry based quantitative assays for proteins linked to PD, AD and HD. Quantitative mass spectrometric assays, like MRM/PRM (multiple reaction monitoring/parallel reaction monitoring), have a huge potential in both the validation of newly discovered biomarker candidates and in the measurements of known markers within a multiplex assay format in the clinical routine. The robustness of the MRM technology has also been reported in inter-laboratory studies presenting high reproducibility that could fulfill the requirements for clinical applications. Recent technological advances make MRM protein assays a convincing alternative to the classical immune-based methods. In addition, mass spectrometry provides sequence information of the targets, i.e., molecular specificity, and has a potential to discriminate and measure different protein variants simultaneously.
The list of our target proteins is the result of a close collaboration with other BAGADILICO groups and data mining in the related literature. We have developed assays for protein markers (inflammatory proteins, apolipoprotein E isoforms) and the assays have been utilized for screening of patient samples both blood plasma and CSF. We are continuously working on new assays and assay extension with an emphasis on the different protein variants (mutations, splice variants, etc.).
The ultimate goal of the study is the application of the established assay panels on samples from large patient cohort, including statistical evaluation of the data; and finally to find correlation between fluid/imaging biomarkers, clinical findings and brain pathology.
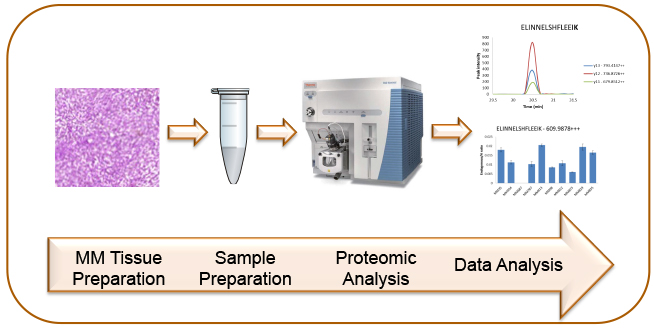
Characterization of malignant melanoma utilizing high resolution MS and bioinformatics.
Malignant melanoma is a disease that is increasing worldwide in fair-skinned populations. In Sweden approximately 2200 new cases are diagnosed every year. At the time of diagnosis 10-15% of these cases are considered advanced and this late discovery of the disease means a very poor prognosis. For malignant melanoma there is today no curative treatment when the tumor has disseminated, because malignant melanoma is relatively resistant to both chemotherapy and radiotherapy. There are either no reliable diagnostic tools that can identify early malignant melanoma and no sensitive methods/tools to follow the progression of the disease, monitor the treatment or identify patients in high and low risk groups, respectively. There is a fast progress within the genetic field, identifying target genes that are associated with the disease (eg. the mutated BRAF gene, V600E). The actual proteins coded and active within disease is the next step in the development of new personalized drugs and targeted diagnosis.
In this project we use both and bottom-up and targeted proteomics strategies (MRM) for proteome profiling in samples from patients with malignant melanoma. The accessibility to the Lund Oncology Biobank with its unique patient samples, both tumor tissues and blood samples, with available clinical data enables us to combine proteome profiling and clinical data. The increased knowledge about protein expression and regulation during melanoma disease progression and treatment will support development of validated biomarkers for melanoma patients.
